

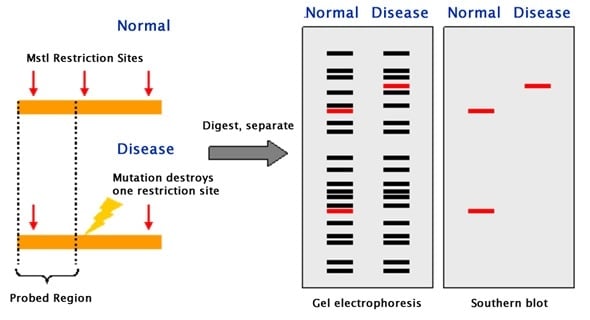
This convenient and simple method is useful in a small basic research study. Terminal restriction fragment length polymorphism (T-RFLP) was introduced to. The mutation is discriminated by the specific restriction endonuclease and is identified by gel electrophoresis followed by staining with ethidium bromide. The PCR-RFLP method allows very rapid, simple, and inexpensive detection of point mutations within the sequences of PCR products. These can be separated by electrophoresis, with the smaller fragments migrating farther than the larger fragments. Here we introduce the PCR-restriction fragment length polymorphism (RFLP) method, which has certain advantages over many other techniques used for analysis of SNPs. Restriction enzymes cut DNA at precise points producing a collection of DNA fragments of precisely defined length. Terminal restriction fragment length polymorphism is a recent molecular approach that can assess subtle genetic differences between strains as well as. Amplified fragment length polymorphism (AFLP) is a PCR-based fingerprinting technique that was first described by Vos et al. Each of these methods has its specific advantages and disadvantages. In conclusion, the presented 16S rRNA gene T-RFLP method is a highly robust. SNP detection is performed by many methods, including hybridization, allele-specific polymerase chain reaction (PCR), primer extension, oligonucleotide ligation, direct DNA sequencing, and endonuclease cleavage. This is usually not achieved by current high throughput sequencing protocols. Progress in single nucleotide polymorphism (SNP) detection technologies has provided information for SNP-based studies, such as identification of candidate genes for the complex genetic diseases, pharmacogenetic analysis, drug development, population genetics, evolutionary studies, and forensic investigations. Restriction fragment length polymorphism (RFLP) analysis was one of the first techniques to be widely used for detecting variation at the DNA sequence level.


 0 kommentar(er)
0 kommentar(er)
